Innovative serotonin sensor to improve neurological drug development
Technological breakthrough provides insights on sleep/wake patterns, social interactions and fear conditioning
A research team led by UC Davis scientists used machine learning to engineer iSeroSnFR, a highly sensitive and selective sensor for serotonin, a neuromodulator that regulates mood and feelings of well-being and happiness. In a study, published Dec. 23 in Cell, the researchers developed and used the innovative sensor to track serotonin in the brains of mice during social interactions, fearful situations and sleep/wake conditions.
The following is a Q&A with lead authors Elizabeth Unger, postdoctoral scholar at the Center for Neuroscience at UC Davis, and Lin Tian, associate professor of biochemistry and molecular medicine at UC Davis Health.
Q: What is serotonin and why is it important?
Unger: Serotonin is a hormone that impacts the entire body and helps with sleeping, eating and digestion. It enables brain cells and other nervous system cells to communicate with each other. Serotonin is linked to many neurological and psychological disorders, including depression and anxiety.
Q: Can you explain the significance of the iSeroSnFR sensor in drug development, especially for neurological disorders?
Unger: Serotonin and its transporters and receptors are the target of hundreds of drugs. Since people respond differently to various drugs, they often try many drugs and doses before they find what works well for them. This search process may take years while the patient suffers.
To develop therapeutic drugs, we must understand the dynamics of serotonin release over time. This is important to deciphering how the complex signals integrate to shape the function and structure of neural circuits. To overcome this challenge, we developed a serotonin sensor, iSeroSnFR, that literally lights up in the presence of serotonin. We can encode this sensor into a virus and either use it in a petri dish or inject it into specific brain regions to reveal serotonin dynamics with high resolution, as triggered by animal behavior. By using the sensor in a petri dish, we created a means of testing different drugs in a lab before giving them to a patient. This allows for better personalized medicine.
Tian: Selective serotonin reuptake inhibitors (SSRIs) are drugs, usually given as antidepressants, that directly target the serotonin transporter. The transporter regulates the amount of serotonin in the brain. A patient may have mutations in the transporter, which could change how it interacts with SSRIs.
In collaboration with the Looger Lab at HHMI Janelia, we developed an assay, or test, to allow clinicians to study the mutations in each patient’s transporter in the lab. The assay is a way to screen many different drugs and determine the efficacy and kinetics of each drug for each patient. This would help guide in choosing a drug and a dose, and hopefully reduce the time needed to start seeing an improvement in patient’s symptoms.
Q: Your experiments looked at the dynamics of serotonin in the central nervous system, specifically the brain. How do you envision it being used in other places?
Tian: Only 5% of all the serotonin in the body is produced in the brain. Most serotonin is produced in the gut, and receptors for serotonin can be found nearly everywhere in the body. On top of that, we know that serotonin can affect many processes such as gut motility and immune function. The good news is that our sensor can be used in any cell type, anywhere in the body. The readout is intensity-based, so any lab with a fluorescent microscope or imaging setup will be able to use it. It’s also genetically encoded, which means that if scientists want to study the gut, they can have the gut cells themselves produce the sensor, rather than trying to work out some sort of complicated delivery method.
Finally, our sensor is soluble, so it can be targeted to any part within the cell. If you want to know about serotonin entering the endoplasmic reticulum, or the nucleus, you can add a little extra sequence to tell the sensor where to go and it will go there. While we didn’t personally do any studies outside the brain, these studies are definitely possible, and we are really excited to hear about the insights that come when the rest of the scientific community uses our sensor for applications that we haven’t even thought of.
Q: What else did you learn? What were some of the more exciting findings in the study?
Unger: The other exciting thing that we did was create a pipeline for developing other tools in the future. Normally molecules like serotonin fit into their binding proteins like a lock and key, with serotonin being the key and its receptor or transporter being the lock. We wanted to engineer a sensor that would bind serotonin, but we didn’t have the right binding partner yet, so we essentially had to create the “lock” out of nothing. Normally this is achieved over millions of years of evolution through the process of natural selection, but we can speed that up using artificial selection that we apply in the lab. We call that directed evolution.
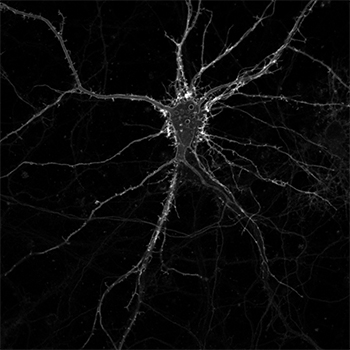
A cultured neuron expressing membrane-bound iSeroSnFR. Credit: Jason Dong
Tian: Liz, who was a postdoc in my lab, pioneered a method using machine learning to speed up the process of directed evolution to radically change the specificity of the binding pocket. In collaboration with UC Davis DataLab and Temple Lang’s group, we spent quite a bit of time developing our method, but in the end it allowed us to make big enough changes to our sensor to create a really good “lock” that fits the serotonin “key” really well. We hope this strategy can be universally applied to binding pocket design and to streamline the sensor development process for any neurochemicals in the brain.
Proteins, for example, can be powerful tools. Every new technique that we add to the protein engineering toolbox makes it easier to create proteins that do exactly what we want. Most engineering efforts tend to add only one or a few mutations to a protein, whereas we ended up adding 19 in total, and actually checked almost 70 different mutation sites. This is a technique that could potentially be used to create other sensors in the future, or indeed, an enzyme that fixes carbon or cleans up oil spills or mass produces spider silk.
Q: How do serotonin levels change based on the state of wakefulness and the sleep-wake cycle?
Unger: We know that serotonin is higher when you’re awake, and lower when you’re asleep. What we didn’t know was the precise timing of that fluctuation. It could be that serotonin is the trigger that causes you to wake up, or it could be that serotonin rises gradually after you’re already awake.
Michael Altermatt in Viviana Gradinaru’s lab at the California Institute of Technology used our sensor to show that serotonin rises within the first few seconds after you wake up. That means it’s probably not the trigger, but definitely closely linked to the awake state. We also found that there is a big difference between REM sleep and non-REM sleep. During non-REM sleep, serotonin is lower than when you’re awake, but when you transition to REM sleep it drops even further within the first few seconds after transition.
There are a bunch of fluctuations during the awake period as well. We don’t know if these jumps are linked to walking, eating or something else, but clearly serotonin is doing something during these times. It is an area that will be studied in the future.
Q: You tested the sensor in mice during social interactions and during fear-inducing situations. What did you find?
Unger: The main purpose of the social interaction and fear conditioning experiments was to demonstrate that the sensor was capable of reporting serotonin dynamics in awake animals behaving normally. Besides successfully proving that, we got some very interesting results.
Serotonin within the brain originates in a small region of the brainstem and is transported to the rest of the brain. Olivia Hon in Thomas Kash’s lab collected data simultaneously from three different brain regions. She found that serotonin is not transported equally in the brain. When mice were allowed to interact with each other, there was a large increase in serotonin in their frontal cortex (known for high level processing) and in the basolateral amygdala (typically associated with fear responses), but not in the stria terminalis (involved in other social interactions).
Fear conditioning is also a complex but very reproducible behavior seen in a variety of animals. When animals expect unpleasant events, they typically respond by freezing in place, especially when unable to run away. During this anticipation time, a lot is going on in their brain to prepare them for this impending experience. The sensor showed that serotonin rises sharply during this freezing period and more so in the frontal cortex than in the basolateral amygdala. After the threat is over, serotonin slowly returns to normal.
The fact that we see increases in serotonin in both social interaction and fear conditioning may not be directly connected. We still have no idea what effect an increase or decrease in serotonin has on the frontal cortex or the amygdala. We also want to be very careful in how we interpret these experiments. Mice have a different social structure than humans and fear different types of things. What we can say is that serotonin is definitely involved in both of these behaviors. Further study in both areas may yield useful insights into human behavior and better interventions for conditions like social anxiety, PTSD or other social and fear responses.
Q: How would you describe your collaboration with other researchers on this work?
Tian: This work is the fruit of an outstanding collaboration among scientists from different fields and departments. We at UC Davis are fortunate to have solid expertise and multidisciplinary collaborations across departments. The collaboration with Vladimir Yarov-Yarovoy’s lab expanded the use of Rosetta to sensor design and with Andrew Fisher allowed us to solve the crystal structure of the sensor for a better understanding of how sensors work.
Liz is a very-gifted scientist who is fearless in executing this large-scale study with rigor and creativity and leading the collaboration to streamline sensor development to characterization. As tool developers ourselves, we rely heavily on collaborations, particularly with those who will be the primary users of our tools. We need their feedback to tailor our tools for their needs. We are accustomed to partnering with different labs across the country and around the world, and these partnerships have been extremely positive and fruitful.
Unger: Our partners at other institutions also bring a different perspective and skillset, which made the whole project come together. We were pleasantly surprised just how enthusiastic people were about the project when we approached them. Everyone jumped on it and went out of their way to provide us with the support we needed. As a postdoc, I am so fortunate to have had such a wonderful experience and enriching training working on this project.
Article: Unger et al. (2020). Directed Evolution of a Selective and Sensitive Serotonin Sensor via Machine Learning. Cell, doi.org/10.1016/j.cell.2020.11.040