Protein Structure and Dynamics Core
About the Facility
Knowing the structural dynamics of the proteins and biological membranes is key to understanding their mechanism of function. Most prominent methods for structural determination are X-ray crystallography and Nuclear Magnetic Resonance Spectroscopy. However crystallized samples select for one static conformation and are difficult to obtain for biomembranes or macromolecular assemblies (such as filaments). NMR methods are difficult for larger biomolecules and membranes, and also carry large sample demands, which may be difficult to obtain.
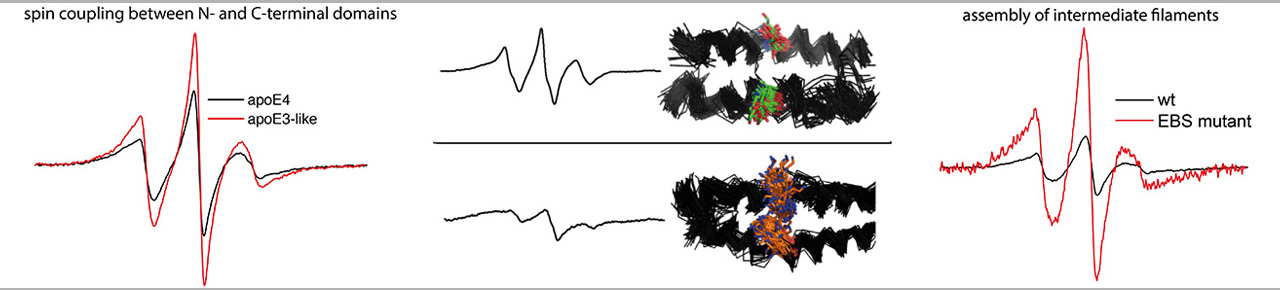
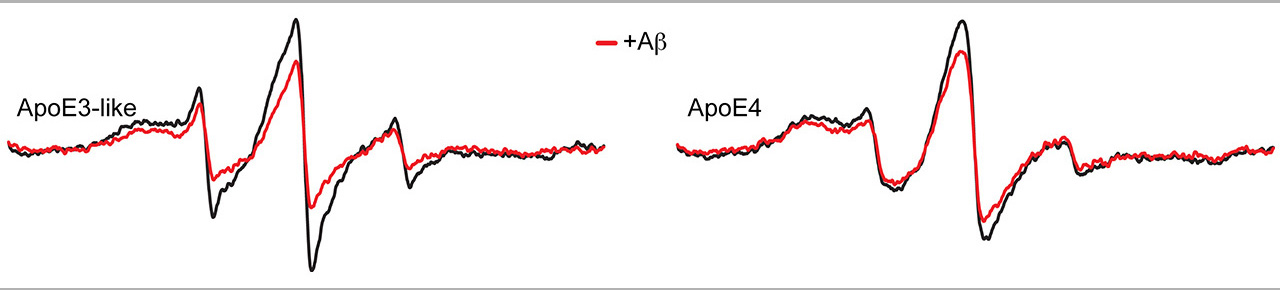
The Site Directed Spin Labeling Electron Paramagnetic Resonance (SDSL-EPR) spectroscopy offers a unique approach to elucidate the structure and dynamics of biomolecules in solution. The approach combines protein chemistry/molecular genetics to target spin probes into biomolecules (most commonly peptides and proteins) with EPR analysis using modifications that can accommodate small biological samples under a variety of conditions (pure protein, tissue, live cells, etc.).
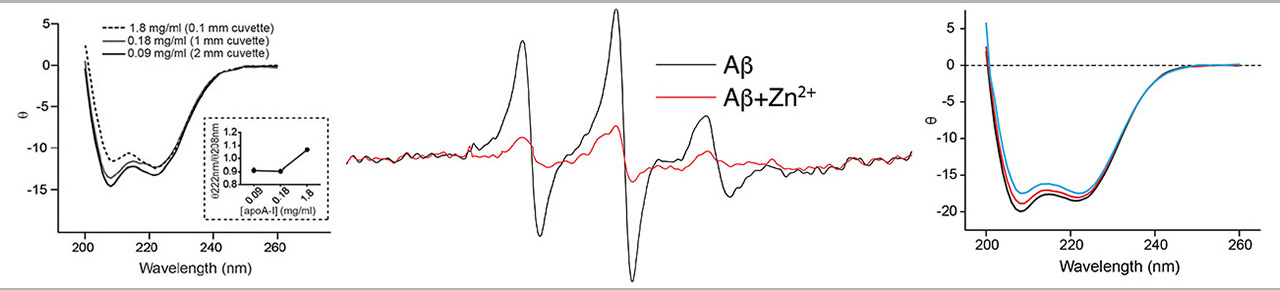
The Circular Dichroism (CD) spectroscopy is the most common approach for studying the overall conformation of biopolymers, such as proteins and peptides. CD is a spectroscopic method based on the interaction of chiral centers in the polymer with circular polarized light, which is very sensitive to levels of regular order in the polymer backbone. CD offers a convenient method for identifying changes in the structure of proteins and peptides.
The EPR spectroscopy and CD spectroscopy is a unique combination of physical methodology used for determining the structural dynamics and molecular structure of proteins and other biomolecules. Such information is crucial for understanding the mechanism of biomolecules and for identifying drugs and other agents that can modulate the behavior of biomolecules involved in disease. The ability to measure structure and dynamics is also important for bioengineering efforts, such as designing platforms for molecular sensing and drug delivery.