Computational Biomedicine
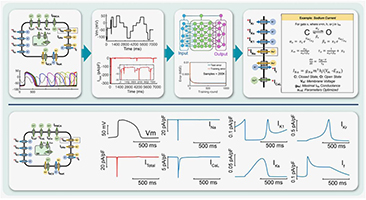
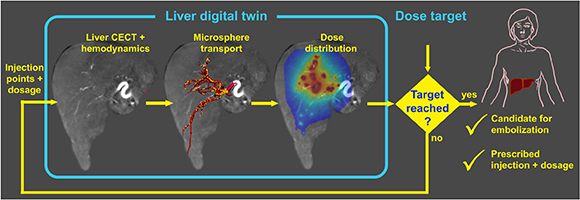
Digital twins are computational analogs of physical systems that continuously evolve through data exchange. They are transforming the relationship between experimentation, computation, and clinical translation and are emerging as a powerful tool for precision and personalized medicine. While established in industrial environments, translating digital twins to biomedical applications faces unique challenges, including inter-individual variability, measurement uncertainty, personalization, and ethical considerations. Credible biomedical twins require solid mechanistic foundations such as electrophysiological, hemodynamic, biochemical, or radiation physics models capturing the spatial and temporal multiscale complexity of biological systems and medical interventions. Hybrid strategies that integrate mechanistic and data‑driven frameworks are most promising to develop accelerated, high‑fidelity surrogates for computationally intensive tasks. Of note, physics‑informed neural networks (PINNs) can embed physics laws directly into the network training process, constraining the model behavior and are rapidly developing for biomedical applications.
Robust deployment of biomedical digital twins will demand rigorous verification, validation, and uncertainty quantification. Establishing unified methodological standards, transparent reporting of model scope and limitations, and federated development will be essential to realize clinically actionable, trustworthy biomedical digital twins.